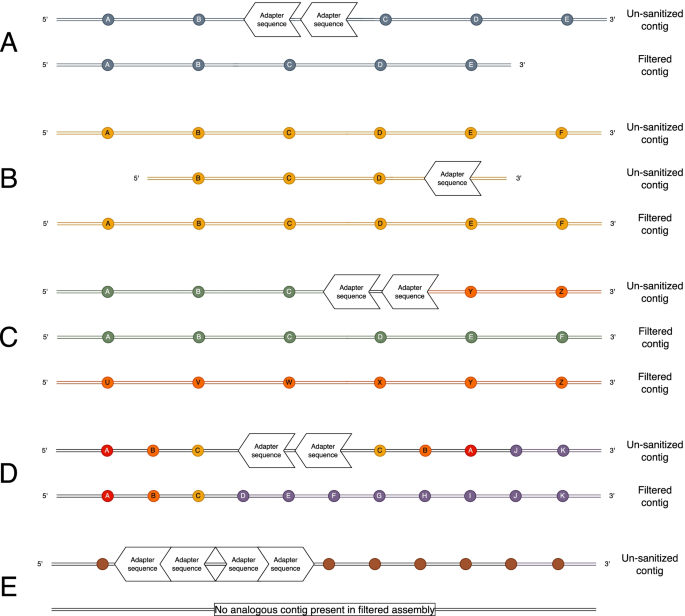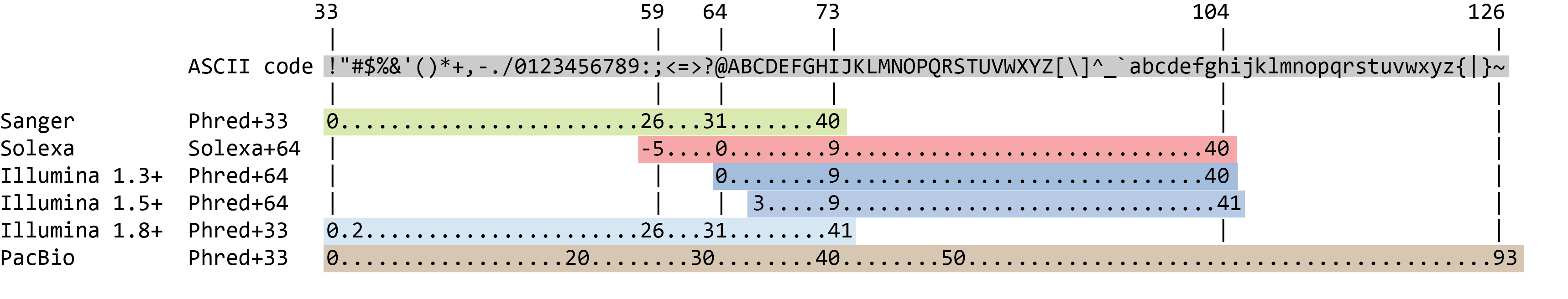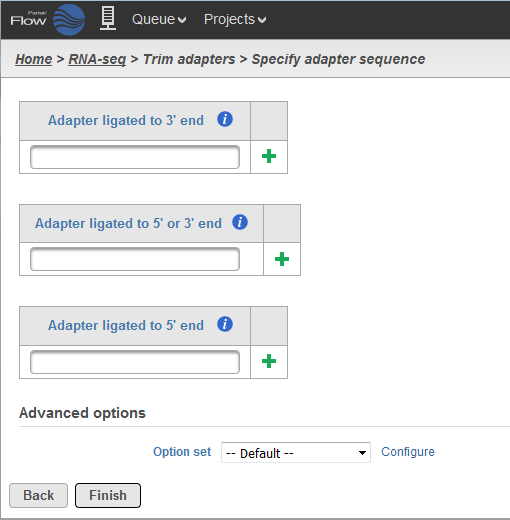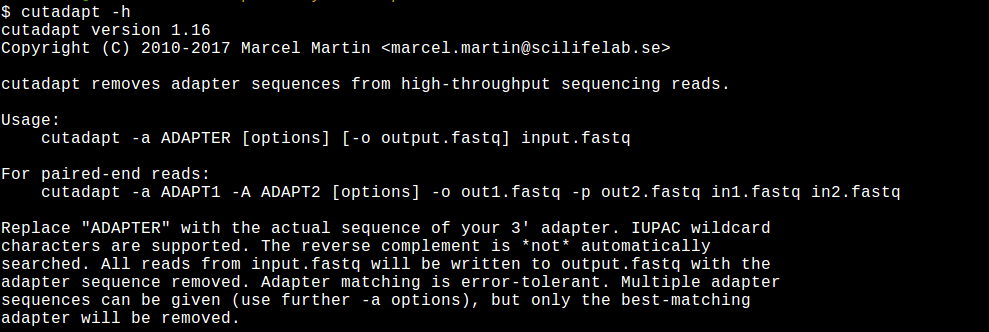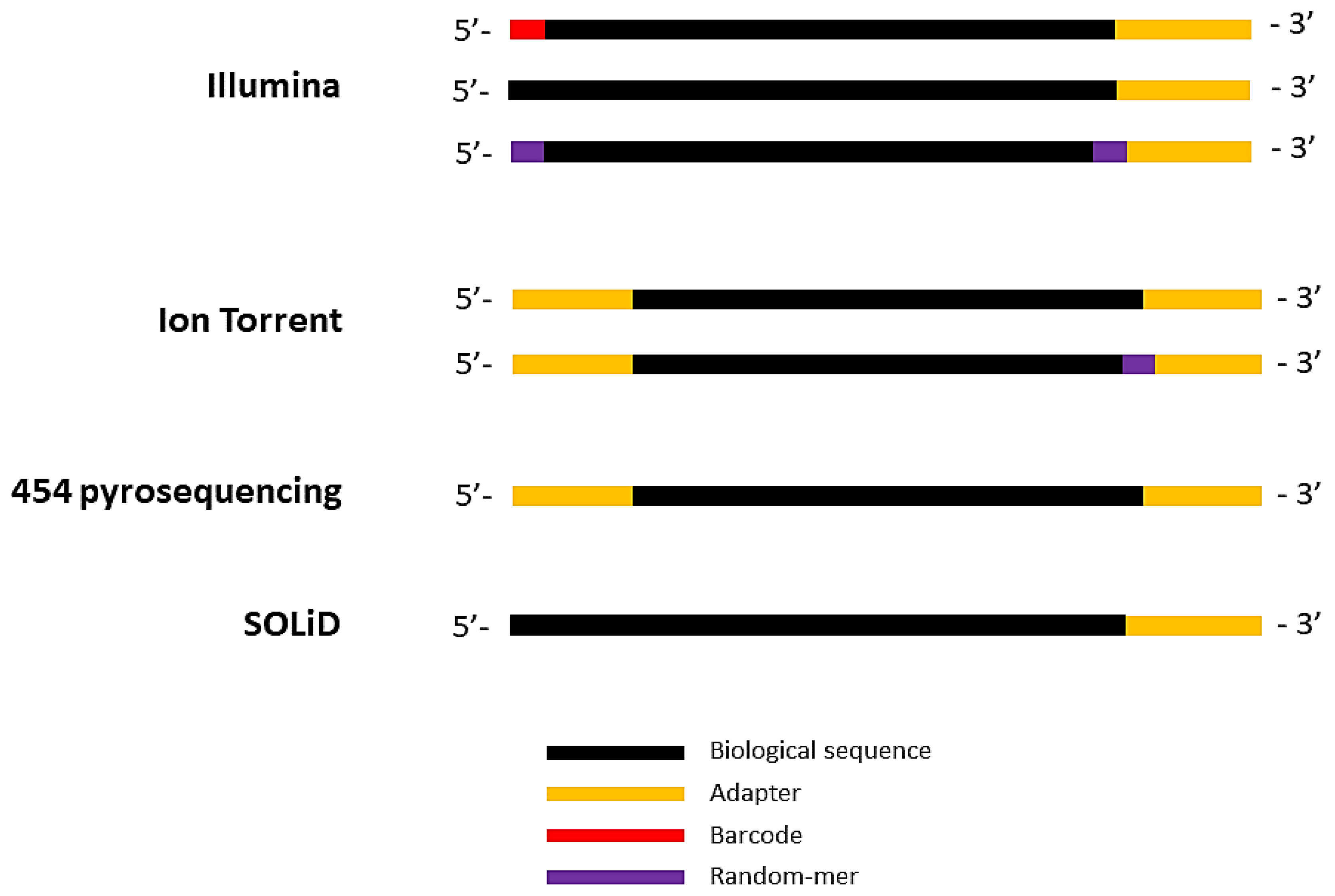
Biomolecules | Free Full-Text | High-Throughput Identification of Adapters in Single-Read Sequencing Data
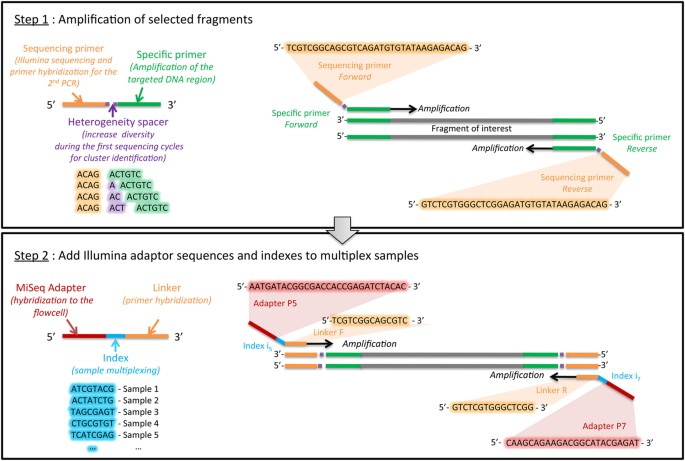
High-throughput sequencing of multiple amplicons for barcoding and integrative taxonomy | Scientific Reports

Kraken: A set of tools for quality control and analysis of high-throughput sequence data☆ | Semantic Scholar

FASTAptameR 2.0: A web tool for combinatorial sequence selections: Molecular Therapy - Nucleic Acids

Kraken: A set of tools for quality control and analysis of high-throughput sequence data☆ | Semantic Scholar
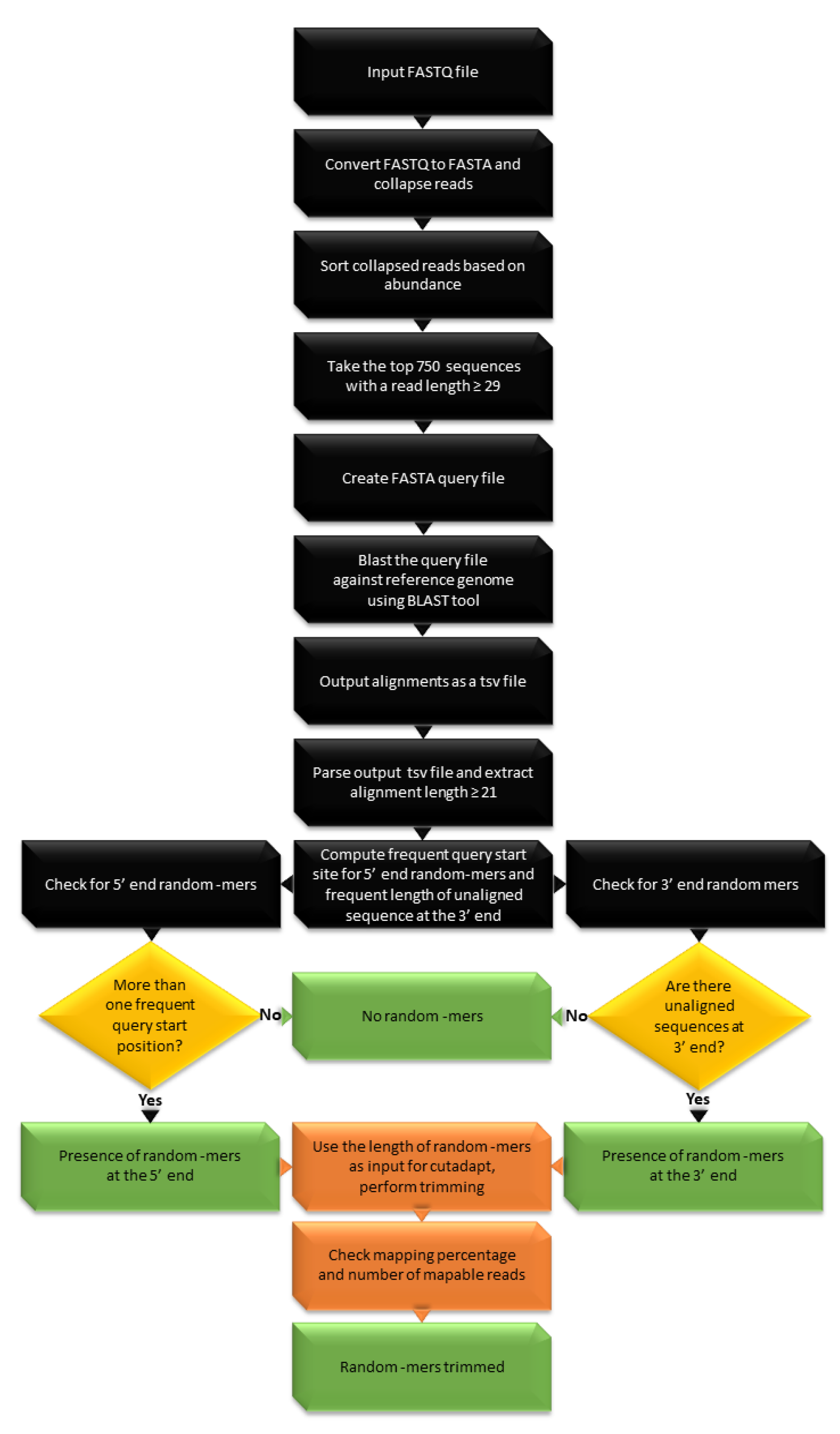
Biomolecules | Free Full-Text | High-Throughput Identification of Adapters in Single-Read Sequencing Data

De-novo Bacterial draft genome de-novo asembly, from the sequencing machine (Illumina) to a genome database (NCBI) An example case: Assembly of Stenotrophomonas. - ppt download
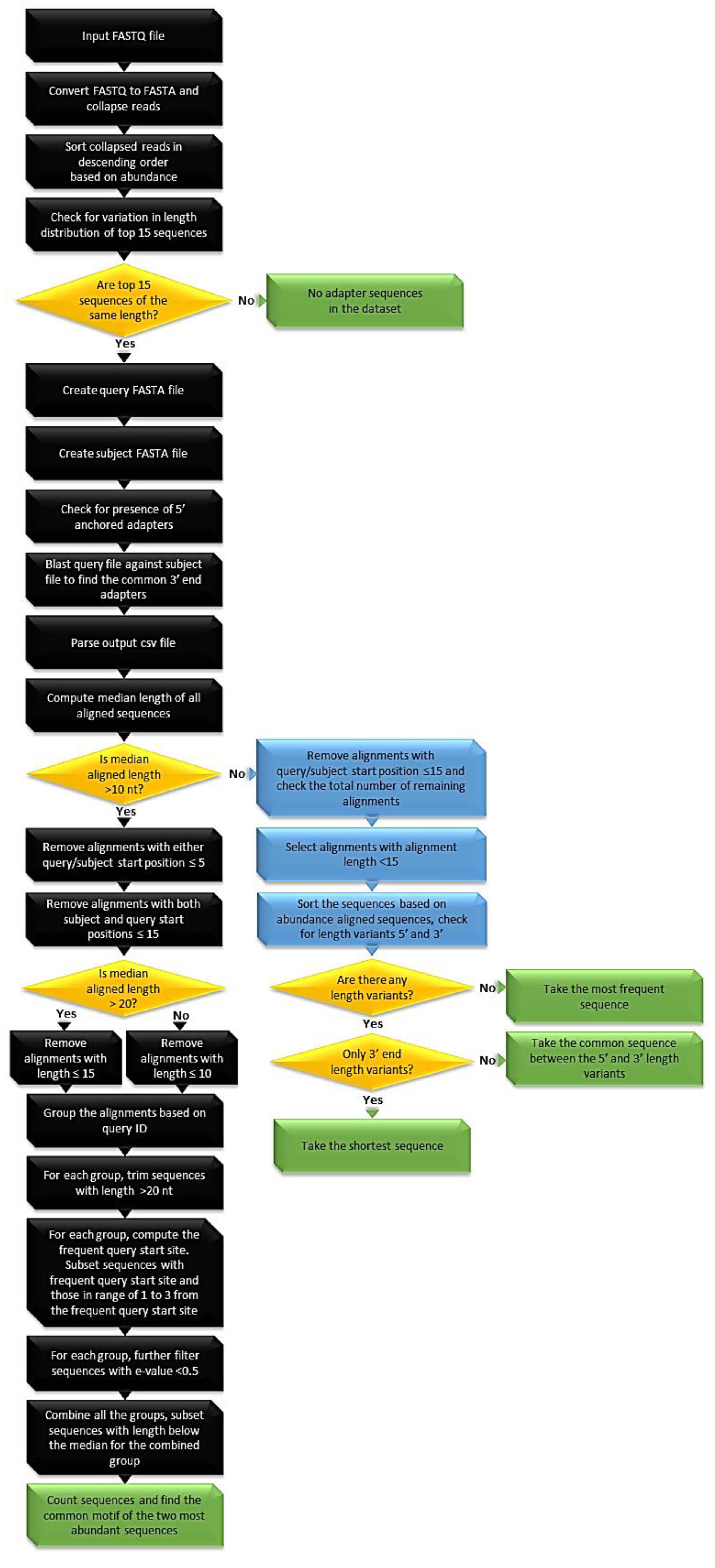
Biomolecules | Free Full-Text | High-Throughput Identification of Adapters in Single-Read Sequencing Data

Handling of targeted amplicon sequencing data focusing on index hopping and demultiplexing using a nested metabarcoding approach in ecology | Scientific Reports
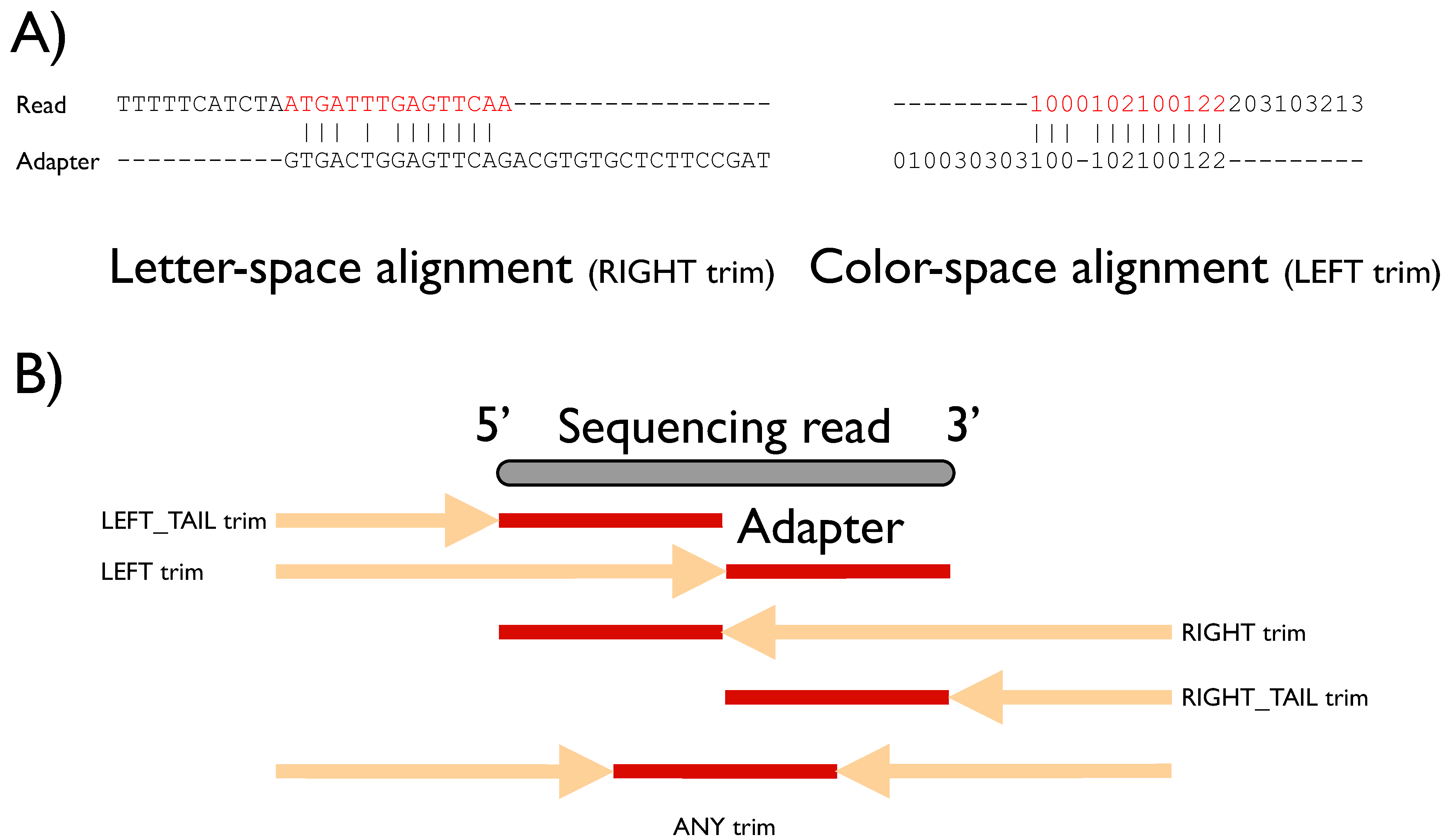
Biology | Free Full-Text | FLEXBAR—Flexible Barcode and Adapter Processing for Next-Generation Sequencing Platforms
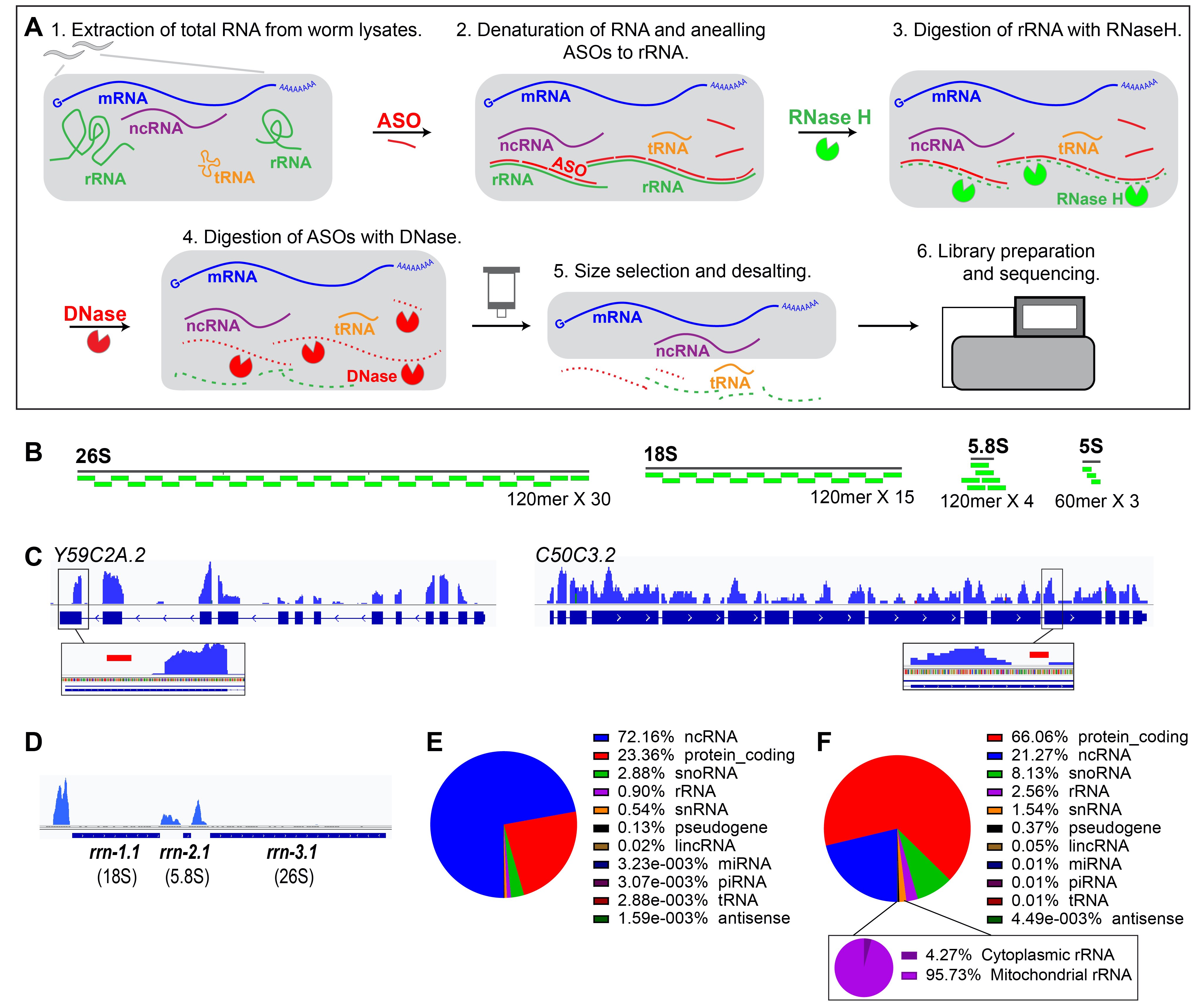
RNA-seq with RNase H-based ribosomal RNA depletion specifically designed for C. elegans | microPublication